For work, I’ve been raising families of RNN’s.
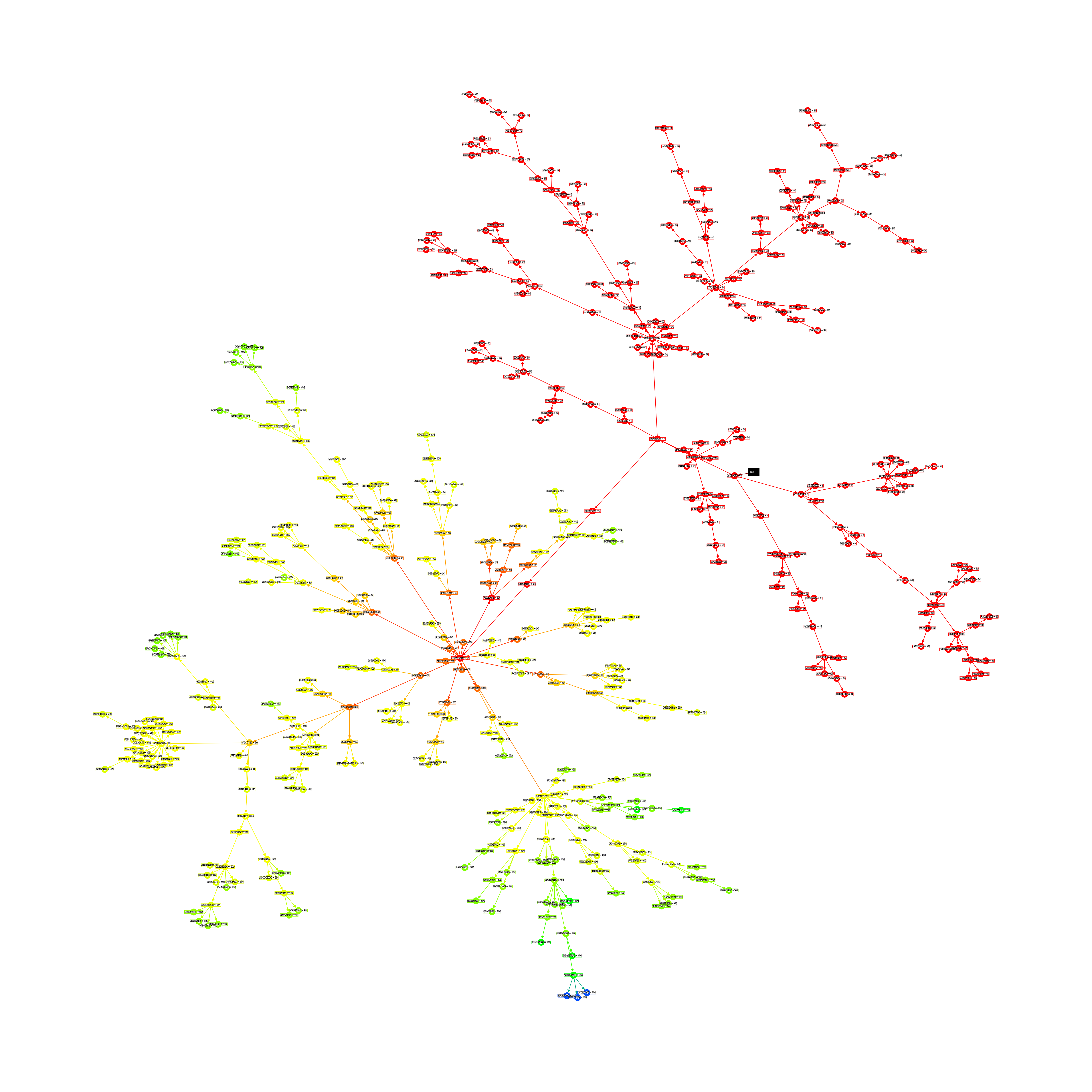
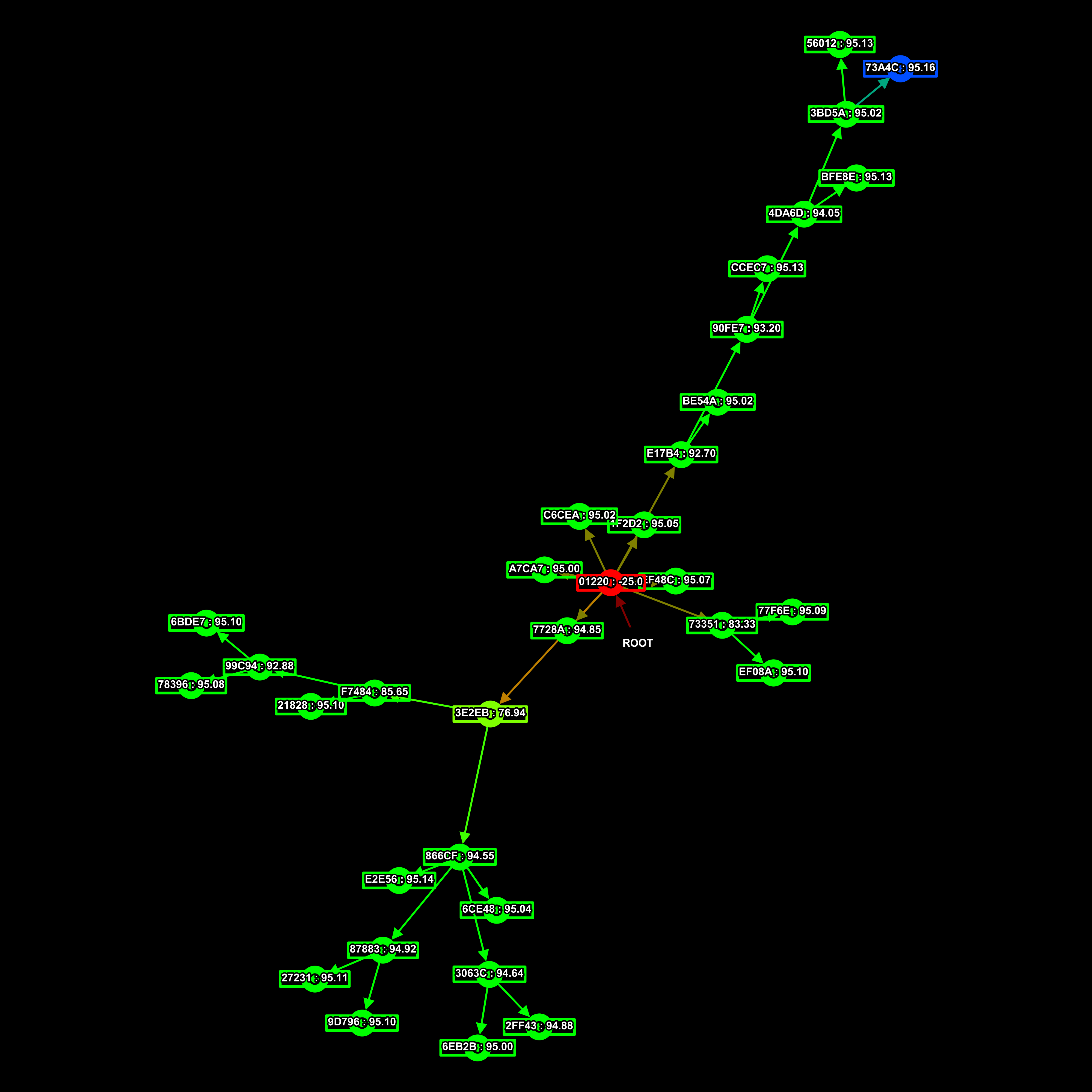
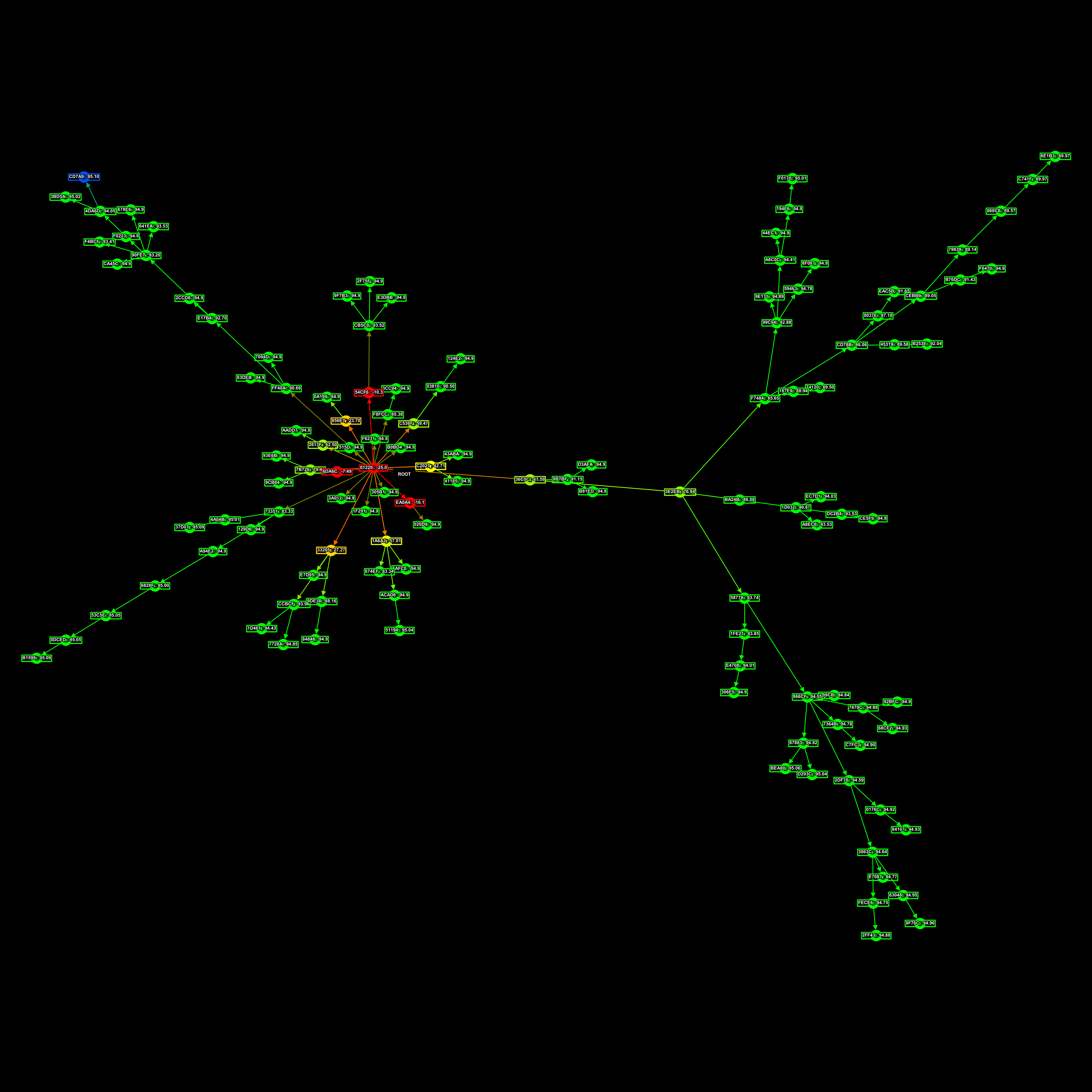
This here is a family-tree of connectomes, AIs. Each point in the graph is an AI with a UUID and a parent. The black rect is the root connectome from which all the others descend.
The fitness of each connectome is represented in relation to all existing connectomes in the tree, by their colour. Red is a low fitness score, yellow intermediate, green high, and blue is the best.
The AIs here are all recursive neural nets, based roughly off the N.E.A.T. paper. We have some modifications to vanilla NEAT, though our design, NEAT6, can be summarised as a recursive neural net able to mutate by:
- Creating connections between neurons
- Deactivating connections between neurons
- Adjusting the weight of any given connection
- Adjusting the number of “steps” to run itself for, before we read the output neurons
- To add new neurons to itself
This is quite different from more traditional, feed-forward NN’s in that connections can emerge that loop in on themselves. This is useful though, since it allows the possibility of long-short-term-memory to arise, letting the AI retain information for longer periods than traditional NN’s. In theory, anyway.
By starting with a very minimal connectome of just the input neurons, a small hidden-layer of maybe as many neurons as there are input neurons, and then the output neurons, NEAT NN’s can mutate into functional connectomes with significantly less connections than other models.
Smaller connectomes are important since all this is running on the CPU, not GPU.
All these AI have the same task: to recreate the first chapter of Harry Potter and The Philosopher’s Stone. We haven’t gotten there yet, but we’ve got some interesting results and came up with some useful methods to raise AI families.
I’d generate a batch of a hundred or so connectomes with the same topology but randomised weights, then start several threads. A thread would select a connectome in the tree and run n trials, a trial being mutating the selected connectome in one of the five previously defined ways that NEAT6 could mutate. After mutation, the thread would run the fitness function on the mutated connectome and, if it scored higher than the unmutated connectome, save and select the mutated connectome for further trials.
We initially only had two types of threads running, defined by their selection function:
- A random-selector thread that simply grabbed any connectome in the tree and ran about 8000 trials on it.
- A best-focus thread that would grab a random connectome from the top-n scoring connectomes, usually the top 20, and run 8000 trials on it.
I figured the high number of trials would mean getting stuck on dud connectomes more often, and then for the best-focus threads, we’d end up creating a lot more children of high scoring connectomes, but not very many of them. As in, we’d select a high-scoring connectome, generate some better children, then the descendants of that initial high-scoring connectome would populate a greater and greater fraction of the top-scoring connectomes. This would lead to the best-focus threads often only focusing on one “species” of connectome.
To remedy this, I simply setup an iterator-thread, reduced the number of trials and increased the range of the top-scoring connectome selector.
The iterator-thread would iterate over every connectome in the tree, running trials on connectomes much further up the family tree than the higher-scoring ones. This meant we’d occasionally create new offshoots that lead to more of the strategy-space being explored than simply running with the first somewhat-successful connectome’s strategy.

The result was a higher top-scoring family-tree being generated in roughly 1/20th the time.
We ran into what we called the “space-barrier” though.
The space-barrier was when an AI found a somewhat-optimal strategy of always outputting the most common character in the first chapter of Harry Potter & The Philosopher’s Stone: ” “.
The character at any given index in the text was marginally more likely to be a space than any consonant, vowel or punctuation. We began work on breaking the space-barrier.
To do that, we’d just have to combine two fitness functions: one that simply compared how close the outputted character was to the real-text [1], and one that reduced the connectome’s fitness score the more repetitive it was. It was rare at first, but eventually the fitness functions were tuned right to produce AI that actually output garble!
I should mention how exactly we read and wrote information from/into these AIs.
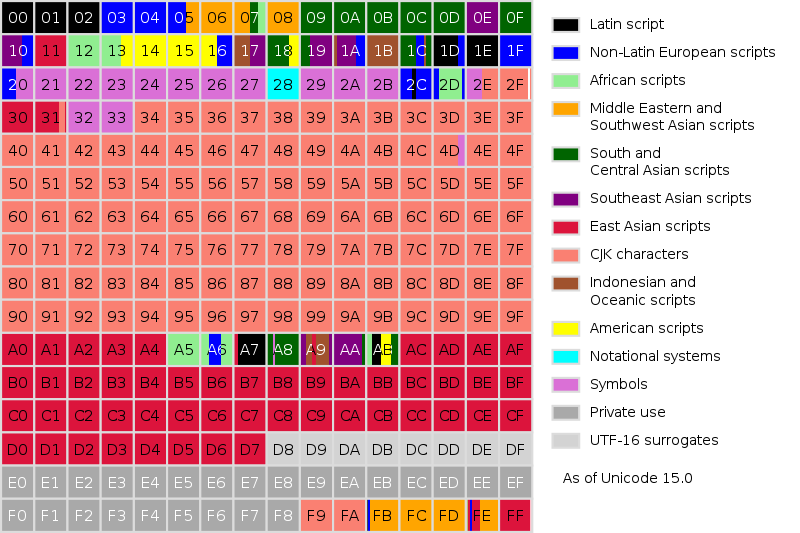
We gave the AIs just enough input neurons to write out a UTF-8 character’s char-code in binary, then enough output neurons for the AI to output enough binary, such that we could read it as a char-code that was then mapped to the appropriate UTF-8 character. We did this so the AI could potentially be multilingual, though later we tried using English tokens.
Now, garble isn’t just garble, there’s degrees to how garbly garble can be. In our case, we could get garble in several different languages, but garble that only used Latin characters was much more valuable than garble in the much larger Asian character set of UTF-8.
It took quite a while, but eventually we got P.
CL-USER> (fitness-func-with-output (fev! (create-network 16 16)))
7.1999974
"PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP"Not long after that, the AI said it’s first word:
CL-USER> (fitness-func-with-output (load-best-ai))
123.70002
" \0c! \0j a a` aa \0i ` \0i aea` a aea \0eaa \0ma e` ea ea ea \0ea a iea \0oa ea a ` \0ma ea \0ma \0i ea \0oa \0k a` ea a \0c! \" \0k \0c! \0oaa` \0k a `a` \" aa \" \0ma a ` a a` \0oa \0ea` \0i \" \" \0j a a` e` ea \0i \0k \0c a \0c \0j ` aea \0k \0k \0a \0o` a a aa a \0oa \0a \0a! a ia \" \0i \0i \0a \0c ea a \0k \0j a a` e` ea \0k! \0a \0k \0a \0i \0ma` \0ma \0oaa` \0c a \0"It said “ma” 🙂
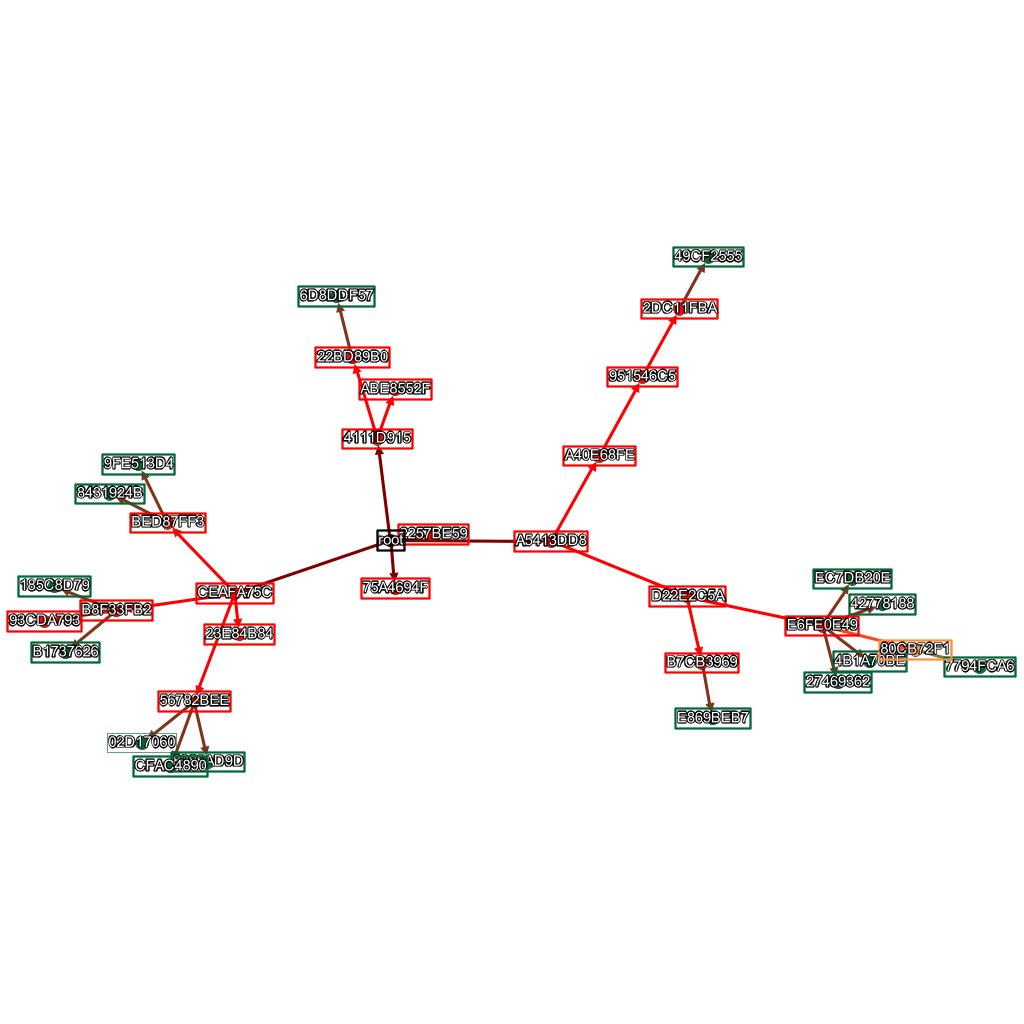
This is the family tree that produced that AI, I don’t know which one it was specifically, but it was one of the blue ones.
It then learned that vowels are much more common than consonants
CL-USER> (fitness-func-with-output (load-best-ai))
106.50003
" \0 d \0\"ee\0\" ee \0ee\0e e e\0egae \0 e e \0\"e e \0e e \0 ee \0 e\0e \0 \0 \0e e \0 ee e \0egea \0 ee\0 e \0e e \0 e d\0\" \0e e \0 \0 e \0 e \0 e d \0 e \0 e\0e \0 eegae \0 e\0 e ee\0e e eea\0ge\0 e e e e \0e e e \0 \0e e \0 ed \0 e \0e \0e e \0egeeaeead\0\"e\0\" ee \0e e\0 \0 e e ge\0ge\0 \0e e \0e \0ge eeaeee \0e e \0 \0 e ed\0\" \0e e\0 \0e e \0e e \0 e\0e \0 e \0 e \0e ee \0 e e \0 \0 \0 e \0 \0e e \0 ee \0 e e e d\0\"ee\0\" ee \0e e\0 e\0 e \0e gea \0 e \0 \0e e \0 e e \0 \0 e \0 e e \0ge\0"And that some consonants are also quite common, like “t” and “w”.
CL-USER> (fitness-func-with-output (load-best-ai))
111.300026
" \0 t \0\"ee\0\" ee \0ue\0u e e\0euae \0\"e e \0\"e e \0e e \0 eu \0 u\0e \0 \0 \0e e \0 ee e \0uwea \0 uu\0 u \0e e \0 e t\0\" \0e e \0 \0 e \0 u \0 u d \0 e \0 u\0e \0 uewae \0 u\0 u ue\0e e uea\0we\0 e e u e \0e e e \0 \0u e \0 ud \0 u \0e \0e e \0uwueaueat\0\"e\0\" ee \0e e\0 \0 e e we\0we\0 \0e e \0e \0ge uuauee \0g e \0 \0 e et\0\" \0e e\0 \0e e \0e e \0 u\0g \0 e \0 u \0u eu \0 u e \0 \0 \0 e \0 \0e e \0 ee \0 u e e t\0\"ee\0\" ee \0e e\0 u\0 u \0e uua \0 u \0 \0u e \0 e e \0 \0 e \0 u u \0we\0"And at another point, it kept greeting us.
CL-USER> (fitness-func-with-output (load-random-ai))
58.10003 : 107F006A-D241-4AD5-840F-88F057881E6E
"hh@Hih\0ihh@Hhhhih뿿i@ih\0ihi`hh@Hihhi@hhihhh@Hhihhi@hhhh\0hhihh\0hi@hh뿿@hihh@hih뿿@hhhh\0hhhhhHhi뿿@iihihii@hihii@뿿ih@hhh뿿@ihhii@hih뿿@hhhh\0hih\0ihhh@hhihih\0뿿ihhH\0H뿿hhhh@hi@@H\0iihiihhH\0ii@Hi뿿hiiih\0hhhhihh\0ih@i뿿hhhhiihhi@@HHHhhh\0hih뿿@ihhh@Hihihh@iiih\0hihi@hhhi@iiihhihhi@ih@Hhhhih뿿@hhh@hih\0Hihhhhih@ih\0H\0Hihi@hHiihH\0Hhhiiiihhi@hiihi@ihHH\0Hhiiihi@ih\0hhh@H\0@ihi@@HHH뿿@ihi@hihi@ihhhi뿿@Hi뿿@ihhii@Hihiihhi@ih\0Hih\0ihhh\0H\0hhh뿿@ihhhh\0iihhhhhihi@ihh@Hhhhih뿿@hhh@hiii@Hih\0@iiihh\0Hih\0ihH\0ihHhi뿿@hhihh\0hih\0hhhhi@Hiihih@ih\0"In an effort to reduce connectome size, we changed the output structure from a binary encoding of a char-code in UTF-8, to just one neuron which would activate according to how confident the AI was that a suggested character was the next character in the text.
We’d pass a series of characters as inputs, for example “Mr. Du”, writing the char-code into the input neurons then stepping the AI and writing the next char-code, to give the AI more information to guess the next character. We also added a feedback neuron that would activate if the AI had guessed the previous character correctly. With all of this, we eventually came to an AI that could say a few select words:

It was saying “and”, “the”, and “Mr Dursley”.
There hasn’t been much progress since Mr Dursley appeared. We ended up running quite a lot of threads concurrently, and had it all networked to synchronise the family-trees across several machines. We setup basic diff-calculation and diff-sharing, wherein any positive mutations found would be saved by calculating the difference between the new and old connectome. We’d then iteratively apply any beneficial diffs to every connectome in the tree, to share useful information across different species. We even implemented “Tipler universes”, where we’d grab a connectome and use it as the root of a new family tree, generate an entire family-tree from it which we could very destructively modify and experiment with, then grab the useful diffs from that tree and exit. The virtual-tree would be destroyed and we’d create a new direct-child of the root connectome in the original tree with all the positive mutations of the virtual-tree. Naturally, we’d then apply those mutations to the entire original tree to search for benefits.
The connectomes’ names were UUIDs, which meant that we could combine different trees together without any name collisions.
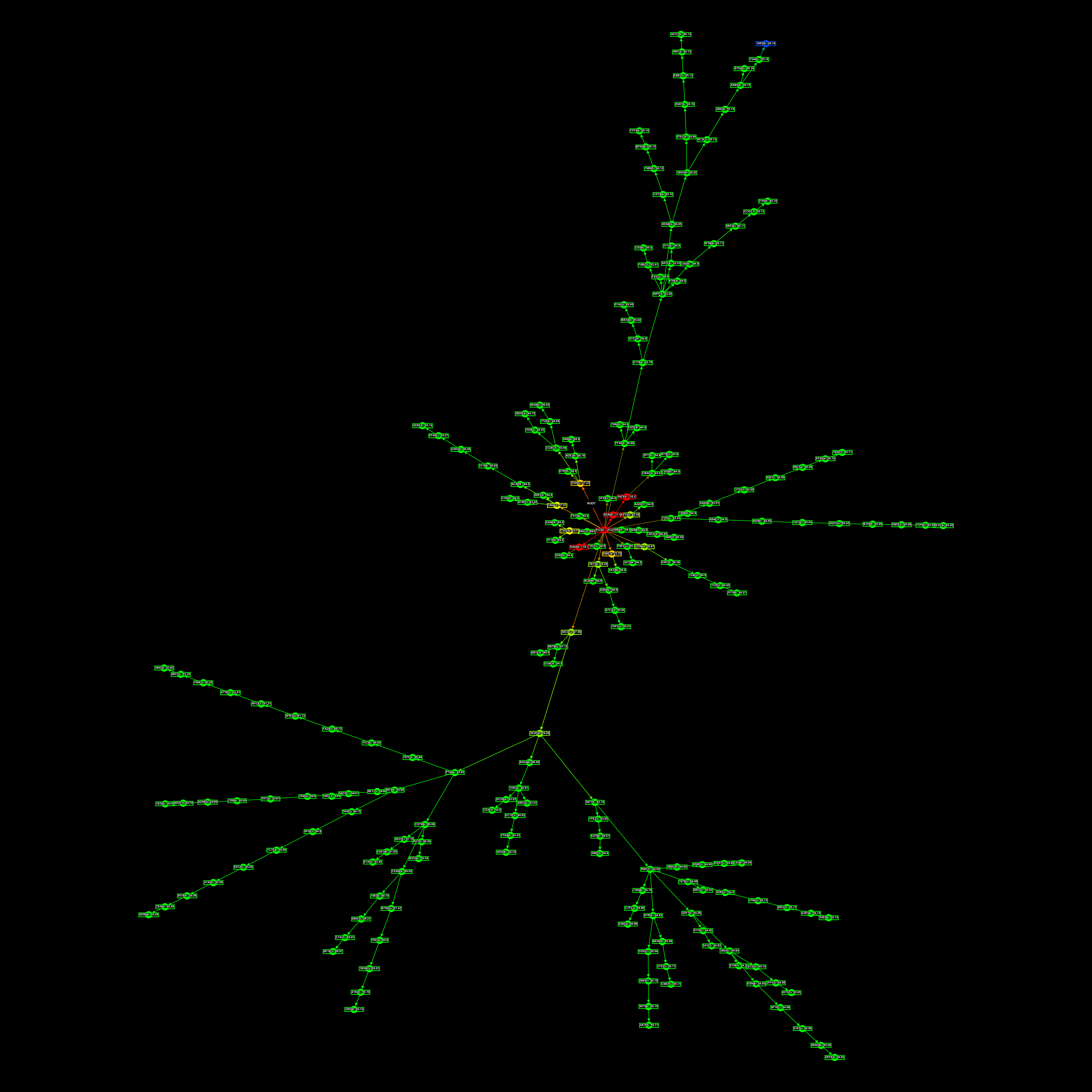
In an effort to reduce tree sizes and thus be able to iterate over a tree faster, I also setup “trimming”, where we’d compress the genetic data of lines of connectomes in a tree, such that connectomes who were only-children and who only had one child were combined. This removed the long spindly lines in the family trees, while preserving all the branches.
Here is the largest family tree I have saved:
And to finish off, here’s an assortment of other RNN family trees I had lying around.






Since all this, I’ve been moved onto a project to create a 2D graphics library for Common Lisp, we’re calling it CLUI.